Genome editing has revolutionized the way scientists can edit DNA sequences in various living organisms. CRISPR, which stands for clustered regularly interspaced short palindromic repeats, is a fast, reliable, and efficient technology for site-directed genome editing compared to early methods such as zinc finger nuclease (ZFNs) or transcription activator-like effector nucleases (TALENS).
CRISPR sequences were first identified as short genomic sequences in bacteria and were discovered to be part of an adaptive defense mechanism. The CRISPR-Cas9 system in E. coli was shown to recognize and cleave complementary DNA sequences, a natural immune process for defending themselves against invading viruses and promoting cellular immunity. The CRISPR-Cas system contains two components: a Cas enzyme and a guide RNA (gRNA), which collectively form the ribonucleoprotein (RNP) complex (Figure 1). The gRNA forms base pairs with a protospacer, which is a target sequence of 20-nucleotides in length. Cas enzymes recognize target sites where cleavage will occur.
When planning your CRISPR experiments, you’ll need to understand the importance of protospacer-adjacent motif (PAM) sequences. In this article, we’ll focus on PAM sequences and considerations you’ll need to know.
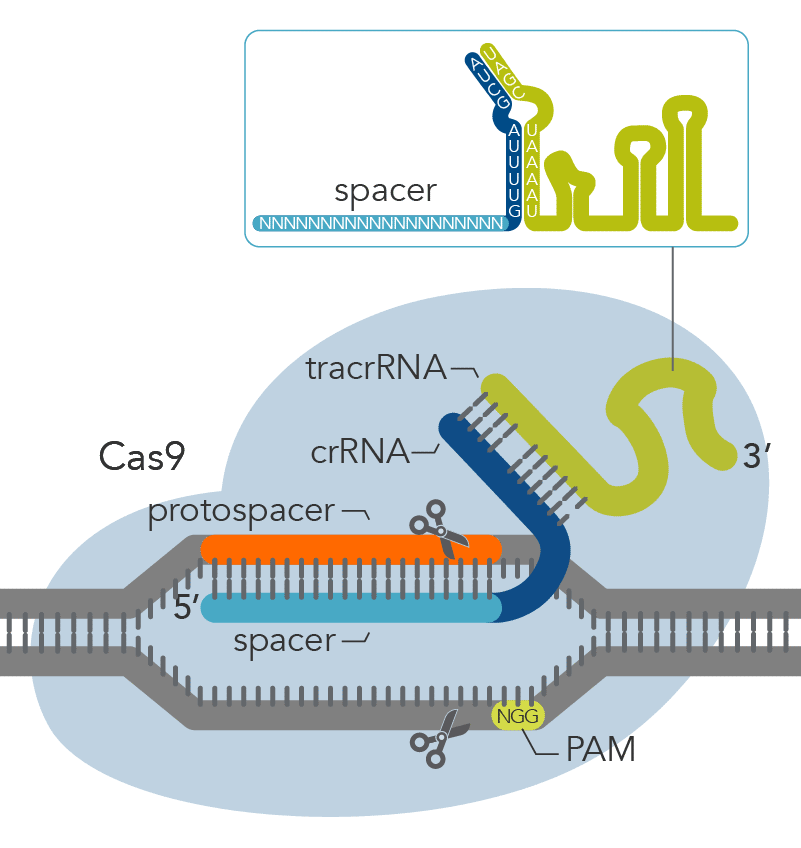
Figure 1. Components of the Alt-R™ CRISPR-Cas9 System for directing Cas9 endonuclease to genomic targets.
What is a PAM sequence?
The protospacer-adjacent motif (PAM) is a short, conserved sequence on the targeted strand of DNA adjacent to the protospacer (the target DNA). The ribonucleoprotein (RNP) complex binds to the protospacer (target DNA) so the Cas enzyme can make a precise cut in the target DNA.
In the invading virus or plasmid, the PAM sequence plays an integral part for distinguishing “self” versus “non-self” and ensures that the CRISPR locus doesn’t get targeted by Cas enzymes and destroyed.
Where is the PAM sequence located?
To be clear, the PAM sequence is not part of the bacterial host genome. The PAM sequence is located next to the protospacer region also referenced to as the target sequence. The Cas enzyme will not be able to bind or cut the target DNA if the PAM sequence isn’t next to the target sequence. Moreover, the gRNA will be able to bind adjacent to the 3’ end of the PAM sequence, which guides the Cas enzyme and tells it where to bind and cut the target DNA.
How does Cas9 recognize the PAM sequence?
The PAM sequence is dependent on which Cas enzyme is used and must be present downstream of the protospacer for cleavage to occur. Depending on the type of Cas enzyme used for gene editing, the PAM sequence will differ. PAM sequences can vary from one bacterial genome to the other. Cas9 proteins derived from S. pyogenes produce the canonical PAM sequence, which is more commonly used in CRISPR experiments.
What is the PAM sequence for Cas9?
For the Sp. Cas9 enzyme, the PAM sequence is a 5’-NGG-3’ sequence, where N is any base. The “N” of the NGG is adjacent to the 3’ base of the non-targeted strand side of the protospacer. For Cas12a V3 or Cas12a Ultra, the PAM sequence is 5’-TTTV-3’ or 5’-TTTN-3’, respectively. The “V” in the TTTV can be one of these nucleotides: A, C, or G. For Cas12a Ultra, a TTTT PAM sequence may work, but may not be as potent.
The role of the PAM site when designing guide RNA
When designing guide RNA for your CRISPR-Cas9 experiment, it is important you do not include the PAM sequence. You shouldn't include the PAM sequence because it will disrupt the gRNA’s function. There is a universal sequence in gRNAs that must follow the protospacer sequence without the PAM being present, or the guide won't function properly (for Cas9). Use this IDT tool to design your Custom CRISPR-Cas9 gRNAs and order your gRNAs here.
IDT offers a complete solution to for your genome editing experiments with the Alt-R™ CRISPR-Cas9 System.