rhAmpSeq™ CRISPR Panels
Highly multiplexed, targeted amplicon sequencing for critical analysis of your CRISPR edits
The rhAmp™ primer pairs in rhAmpSeq CRISPR Panels mitigate primer dimers and maximize multiplexing to provide you with high-quality amplicon libraries. Analyze more samples faster with the confidence of uniform coverage and the time savings of efficient library prep. The rhAmpSeq Design Tool makes it easy to input your targets, and our speedy delivery means you can assess your CRISPR results sooner.
Ordering
With our rhAmpSeq CRISPR Panels you have several advantages:
- Interrogate CRISPR edits in a variety of species with your own custom panel
- Multiplex hundreds of primer pairs in a single pool
- Select from a flexible set of delivery scales and formats
rhAmpSeq CRISPR Panels
Create custom NGS amplicon panels.
To complete your rhAmpSeq CRISPR Analysis System workflow, you will also need the rhAmpSeq CRISPR Library Kit and rhAmpSeq Index Primers. The rhAmpSeq CRISPR Library Kit includes Analysis Credits to enable data processing and quantification of editing events with the rhAmpSeq CRISPR Analysis Tool.
Product Details
rhAmpSeq CRISPR Panels are highly multiplexed primer sets for targeted sequencing. These panels have been expressly designed for a wide range of species. They are Custom rhAmpSeq Panels specifically designed to assess CRISPR edits and are ideal for confirming on- and off-target CRISPR gene editing experiments.
Design custom panels for your genome editing research
Our rhAmpSeq Design Tool lets you quickly and easily design custom rhAmpSeq CRISPR Panels specific to your application and targets of interest (Figure 1). If you would like additional assistance, our expert bioinformatics team can help tailor a design for your custom project (Contact us).
Once your design is complete, we provide a detailed summary report of the custom panel results for you to review and, when necessary, iterate your design before you order your custom rhAmpSeq CRISPR Panel. Also available for download are the associated assay BED files and assay IDs to order any sub-panel from the set of assays that were a part of your custom design.
Custom rhAmpSeq design
Custom assay design involves two key steps that provide improved performance over other amplicon sequencing systems:
- Assay design includes extensive target site review and rhAmp primer design for each target, followed by a comprehensive QC of each rhAmp primer to mitigate off-target effects, such as primer-dimer formation and non-specific genomic hybridization.
- Virtual assay pooling is a unique multiplex primer QC feature that further enhances the on-target amplification of rhAmp PCR technology. It allows for more accurate predictions of which potential primer-dimer amplification products could lower overall reaction efficiency and decrease the percentage of correctly mapped reads.
Any assay that is successfully designed but not able to be pooled into the primary panel will be placed in secondary panels. These secondary panels have a designated prefix (such as P2 or SC) in the resulting panel name that correlates to user input during the design submission. Refer to the rhAmpSeq Design Tool User Guide for more information.
Technical details
Custom rhAmpSeq CRISPR Panels are available in three convenient scales to fit your experimental needs: 0.4 nmol, 4 nmol, and 8 nmol. As shown in Table 1, we recommend adjusting rhAmp primer concentrations in the first amplification reaction (Targeted rhAmp PCR 1) depending on your panel size (plexity). Table 1 shows the approximate number of reactions for each scale based on panel size.
Table 1. Number of rhAmp primer reactions based on scale and panel size.
| Panel size | rhAmp primer concentration in 10X panel* | Number of reactions per rhAmp primer order | Example | |||
|---|---|---|---|---|---|---|
| Panel size | Number of reactions | |||||
| 0.4 nmol primer (x) | 4 nmol primer (x) | 8 nmol primer (x) | ||||
| ≥500-plex | 50 µM total primers | x nmol of primer/(0.1 nmol total/panel size) | 1000 | 4000 | 40,000 | 80,000 |
| 750 | 3000 | 30,000 | 60,000 | |||
| 500 | 2000 | 20,000 | 40,000 | |||
| 101–499-plex | 100 nM each primer | x nmol of primer/0.0002 nmol per rxn | 400 | 2000 | 20,000 | 40,000 |
| 200 | 2000 | 20,000 | 40,000 | |||
| ≤100-plex | 250 nM each primer | x nmol of primer/0.0005 nmol per rxn | 100 | 800 | 8000 | 16,000 |
| 50 | 800 | 8000 | 16,000 | |||
| 25 | 800 | 8000 | 16,000 | |||
* Important: When creating rhAmpSeq primer pools, do not to combine forward and reverse primers for long-term storage. The primer concentrations in the 10X panel stocks assume there will be a separate forward pool of primers and a separate reverse pool of primers. Refer to the protocol in the Resources section for details.
Product Data
Panel uniformity
High uniformity and on-target amplification with rhAmpSeq CRISPR Panels allow for interrogation of on- and off-target editing in a single library prep with PCR amplification as represented below using the popular AAVS1 “safe harbor” site (Figure 2). All 28 empirically identified off-target sites can be verified when Cas9 is constitutively expressed in HEK293 cells. Using ribonucleoprotein (RNP) delivery of the Cas9/guide complex in conjunction with Alt-R™ S.p. HiFi Cas9 Nuclease V3 dramatically reduces off-target editing.
Figure 2. High uniformity and on-target amplification with rhAmpSeq CRISPR Panels allow for interrogation of on- and off-target editing in a single library prep with PCR amplification. HEK293 cells constitutively expressing S.p. Cas9 nuclease were electroporated with 10 µM AAVS1-targeting Alt-R sgRNA. Alternatively, standard HEK293 cells were electroporated with 4 µM Alt-R wild-type (WT) or HiFi Cas9 Nuclease complexed to the AAVS1 sgRNA (at a 1:1.2 protein to gRNA ratio), including 4 µM Alt-R Cas9 Electroporation Enhancer using the Amaxa™ Nucleofector™ 96-well Shuttle™ System (Lonza) (n = 1 transfection per condition). gDNA was isolated and amplified using a custom rhAmpSeq CRISPR Panel containing amplicons for the on-target and 28 of the top off-target sites identified by GUIDE-Seq. Amplicon sequencing on the Illumina MiSeq™ (v2 chemistry, 150 bp paired-end reads) was performed and analyzed using the rhAmpSeq CRISPR Analysis Tool (n = 1 amplification per condition). (A) The histogram of panel coverage shows 100% of assays have read coverage depth that is >0.2X of the mean read coverage depth for all assays in the panel, indicating highly uniform enrichment via amplification. (B) Fragment analysis of final panels in duplicate shows the expected fragment sizes of 300–400 bp with no primer dimers present. (C) NHEJ editing for the on-target locus (Assay 1) and off-target loci (Assays 1–29) is reported using an untreated control for background subtraction. Background editing was <0.2% for all assays in this panel.
Resources
Frequently asked questions
Why is it that my main rhAmpSeq™ CRISPR Panel does not contain assays against all my targets?
The rhAmpSeq Design Tool aims to maximize PCR target specificity at intended loci and minimize any amplification of unintended loci that might consume valuable sequencing reads. Therefore, the design output can sometimes include not only the main panel, but also a secondary pool and possibly individual (singleton) primer sets requiring independent amplification.
Several factors can lead to more than one panel being designed:
- The PCR targets may be too close together. The rhAmpSeq Design Tool requires a minimum distance of 600 base pairs (bp) between targets. Targets that do not meet this minimum distance requirement impede having two unique primer pairs in a single pool. Typically, this results in an adjacent target needing to be covered by either a secondary pool, or as a singleton assay, if the assays cannot be merged.
- The algorithm may not be able to select a primer pair that is both specific enough to cover the PCR target of interest, and distinct enough to allow pooling (multiplexing) without creating primer dimers.
- The ideal primer pair for a given target may amplify a repetitive region. In this case, the optimal primer pair must remain a singleton primer pair to avoid impacting the operation of the primary pool.
- An assay with potential issues is less likely to be pooled due to the risk of impacting the overall panel results. Examples of these types of potential issues include the following:
- A primer overlaps common single nucleotide polymorphisms (SNPs) leading to potential inefficient amplification.
- The GC content of a primer or amplicon significantly differs from the panel’s average GC content.
- The Tm or secondary structure of the primers is not ideal.
- The amplicons contain homopolymers.
- The locus and design constraints do not permit the design of a primer pair that is expected to uniformly amplify with the assays in the primary pool.
How do I design a rhAmpSeq™ CRISPR Panel to evaluate on- and off-target editing using the rhAmpSeq CRISPR Analysis Tool?
For compatibility with the rhAmpSeq CRISPR Analysis Tool, we recommend submitting the targets of interest to the rhAmpSeq Design Tool as a 6-column BED file that includes the following information: chromosome, start, stop, desired name, mismatch # (or a 0), and strand (+ or –).
Additionally, consider the following criteria with the BED file input:
- The BED file needs to be devoid of headers.
- The input to the tool should be the coordinates of your on- and off-target gRNA binding loci (i.e., usually 20–24 bases in length).
- The guides should include “strand” information. This is needed for the rhAmpSeq CRISPR Analysis Tool to report results accurately. Failing to provide strandedness will result in use of an incorrect, non-optimal editing window in the analysis software.
- Guide locations should NOT include the PAM sequence; remove the PAM sequence from the target location in the BED file. Failure to remove all the PAM information can adversely affect data analysis.
The rhAmpSeq CRISPR Analysis Tool has the following run restrictions:
- Multiplexed amplification pools are limited to 500 target sites per pool.
- Input FASTQ files should be <1 GB in file size.
Only the following organisms are supported by rhAmpSeq CRISPR Analysis Tool:
- Homo sapiens, human (hg38 & hg19)
- Mus musculus, mouse (mm10)
- Caenorhabditis elegans (Wbcel235)
- Rattus norvegicus, rat (rn6)
- Danio rerio, zebrafish (Grcz11)
- Macaca mulatta, rhesus monkey (rheMac8)
- Macaca fascicularis, crab-eating macaque (macFas5)
What is the Analysis Lab?
The Analysis Lab is a platform that IDT created to enable multiple researchers to collaborate and access the rhAmpSeq™ CRISPR Analysis Tool simultaneously, share experimental findings, and manage rhAmpSeq Analysis Credits.
Activation codes are redeemed, added to the platform, and then can be used to analyze data with the rhAmpSeq CRISPR Analysis Tool.
What is the analysis platform that powers the rhAmpSeq™ CRISPR Analysis Tool?
CRISPRAltRations is the analysis platform behind the rhAmpSeq CRISPR Analysis Tool.
This IDT-developed analysis pipeline is used for CRISPR on- and off-target editing analysis of next generation sequencing (NGS) data generated from amplicon sequencing. The analysis platform available through the rhAmpSeq CRISPR Analysis Tool utilizes cloud-hosted computational resources for data processing.
Briefly, the CRISPR workflow is as follows:
1. Read pairs are identified and merged from paired-end sequencing.
2. Reads are binned to the expected amplicons, resulting from targeted amplification library preparation (e.g., the rhAmpSeq CRISPR Library Kit).
3. A Cas-enzyme specific aligner aligns the read to the expected amplicon.
4. Variants are called and summarized.
Although this workflow is relatively common to most software tools that analyze NGS data derived from CRISPR screens, the rhAmpSeq CRISPR Analysis Tool has key improvements that help to enable more correct variant identification, including: a Cas-specific aligner; an optimized default variant identification window; systematically substantiated program parameters that use open source tools to provide high-quality results.
Do IDT's rhAmpSeq™ Analysis Credits expire?
Yes, your rhAmpSeq Analysis Credits expire after 1 year.
IDT's rhAmpSeq Analysis Credits are automatically redeemed from the earliest expiration date to the latest.
View the expiration dates on your rhAmpSeq Analysis Credits in the Analysis Credit balance details window found in the Analysis Lab.
What is the recommended sequencing depth to quantify CRISPR editing?
A target coverage of 1000 paired end reads per amplicon is recommended to quantitate CRISPR editing.
What is the sequencing activity of the rhAmpSeq™ system?
For Custom rhAmpSeq Panels, we typically observe >90% mapped reads, >90% on-target rates, and >85% coverage uniformity for human panels sizes up to 3000-plex using highly purified reference gDNA.
Results may vary based on several factors, including species and availability of reference genome, input targets, panel size, and sample type/quality.
How is the concentration of each primer in a rhAmpSeq™ 10X forward or reverse pool calculated for a ≥500-plex panel?
Use the following formula to determine the concentration of each rhAmp primer in the 10X forward or 10X reverse pool:
rhAmpSeq ≥500-plex:
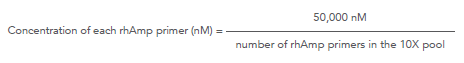
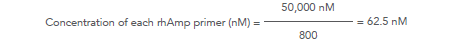
For example, for a 800-plex panel:
What sample types can I use with rhAmpSeq™ assays?
The rhAmpSeq assay is compatible with purified genomic DNA, cell-free DNA (cfDNA), and formalin-fixed, paraffin-embedded (FFPE) tissue.
What are the advantages of the rhAmpSeq™ Library Preparation Protocol and the rhAmpSeq High Throughput Library Preparation Protocol?
The rhAmpSeq™ Library Preparation protocol offers high assay performance (i.e., more mappable reads and better coverage uniformity between assays), while the rhAmpSeq High Throughput Library Preparation Protocol is a streamlined protocol that saves time and overall cost.
Why does the rhAmp primer in my rhAmpSeq™ CRISPR assay contain an N base?
In cases when the best rhAmpSeq CRISPR assay contains a primer overlapping a common SNP, the affected base will be replaced with an N degenerate base.
How much time does each rhAmpSeq™ CRISPR protocol require?
The rhAmpSeq CRISPR Library Preparation Protocol takes approximately 4–4.5 hours with 1–1.5 hours of hands-on time.
Note: Times listed are for processing 12–96 samples using manual pipetting, including reaction setup, cleanup, library quantification, and normalization steps. The time for either protocol will vary depending on the number of samples being processed.
Why did some of my targets not have rhAmpSeq™ assay designs?
The IDT design algorithm generates designs based on a complex set of parameters that predict uniform results.
Some targets contain genomic sequences that prevent us from creating primer pairs within these limits.



