rhAmp™ Genotyping Master Mix and Reporter Mixes
Amplification and reporter mixes optimized for rhAmp SNP Genotyping
rhAmp Genotyping Master Mix and rhAmp Reporter Mixes are specially formulated for use with rhAmp SNP Assays to deliver signal-to-noise ratios for high-confidence genotype calls. rhAmp Genotyping Master Mix is a two-enzyme formulation ideal for precise amplification and allele discrimination. rhAmp Reporter Mixes contain a universal 5' nuclease reporter system to distinguish the reference allele from the alternative allele for any rhAmp SNP Assay.
Ordering
rhAmp Genotyping Master Mix and Reporter Mixes
- Easily report genotype calls with the RNase H2 assay chemistry
- Generate high-confidence calls using a novel Taq polymerase with enhanced allelic discrimination
- Cost-effective universal reporter probes can help save on custom probe costs
- Use with common commercial real-time PCR instruments by selecting mix with or without reference dye
Product Details
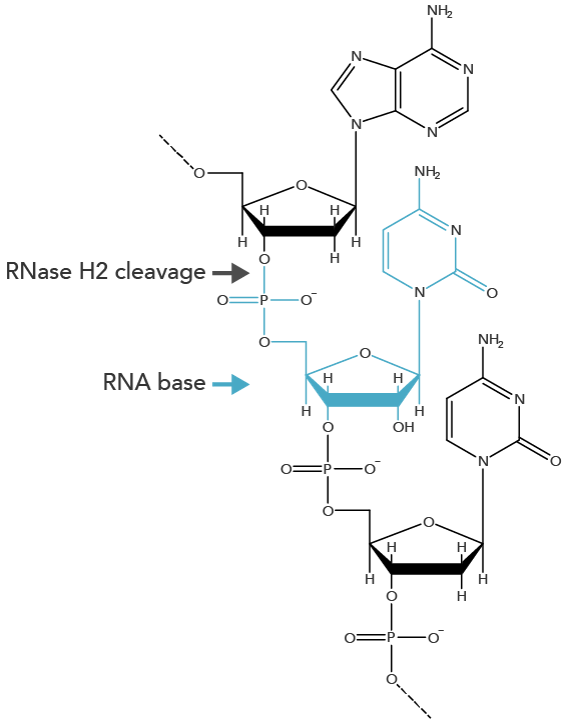
rhAmp SNP Genotyping technology incorporates a novel method using blocked PCR primers that contain a single RNA base. The rhAmp primers are activated when the blocking motif is removed by the RNase H2 in the rhAmp Genotyping Master Mix. Cleavage occurs at the 5' end of the RNA base (Figure 1) only when it is perfectly matched to its DNA complement. Primer cleavage occurs in the background during each anneal/extend cycle to generate a 3'-hydroxyl for polymerase extension, which means the PCR protocol does not require any additional steps. The mechanism creates extremely tight control of amplification that greatly reduces primer-dimers and off-target amplification of closely related sequences.
Figure 1. RNase H2 cleavage produces a 3'-hydroxyl group for extension by a proprietary Taq polymerase. RNase H2 cleaves the phosphate backbone between the DNA base and RNA base during normal PCR cycling conditions. This produces a 3'-hydroxyl group on the DNA base that is accessible to DNA polymerase extension.
rhAmp Genotyping Master Mix
rhAmp Genotyping Master Mix contains hot start versions of RNase H2 from Pyrococcus abyssi (P. abyssi) and a novel Taq DNA Polymerase for enhanced discrimination of allelic mismatches. This dual enzyme formulation, combined with rhAmp SNP Assays, provides high-affinity, precise PCR for the identification of single-nucleotide polymorphisms [1].
P. abyssi is hyperthermophile. Therefore, the P. abyssi RNase H2 enzyme has optimal activity between 70−75°C and is functional in rhPCR between 50−75°C. P. abyssi RNase H2 has very low activity (~1000X less active) at room temperature. These properties mean that the enzyme is ideal for use in the rhAmp Genotyping PCR buffer conditions. The genotyping Taq DNA Polymerase is a novel, proprietary polymerase that is ideal for allelic discrimination. The Taq polymerase and RNase H2 combination delivers clear discrimination and high signal-to-noise ratios, due in part to reagents not being unnecessarily consumed by primer-dimers and spurious amplification products.
rhAmp Reporter Mix
rhAmp Reporter Mix is a cost-effective, 5′ nuclease assay reporter mix that contains 2 universal probes for genotyping. The FAM universal probe is assigned to the reference allele, and the Yakima Yellow® universal probe is assigned to the alternate
allele. The Yakima Yellow reporter dye can be read on qPCR instruments in the VIC® channel without need for calibration.
The universal primer and probe designs have been made to deliver consistent results for high-quality automated
allele calling. And, unlike traditional 5' nuclease SNP assays, the rhAmp SNP universal reporter system saves money because you won’t need to synthesize 2 SNP-specific probes for every assay. This system helps to enable rhAmp SNP Assays to generate
high-quality genotype data (Figures 1−3).
- rhAmp Reporter Mix sizes are configured to complement rhAmp Genotyping Master Mix product sizes for easy planning and ordering (Table 1)
- Available with or without reference dye for maximum flexibility on all qPCR platforms (Table 2)
Table 1. rhAmp Reporter Mix, with or without reference dye, matched to correct size rhAmp Genotyping Master Mix.
| rhAmp Reporter Mix* or rhAmp Reporter Mix w/reference dye† | rhAmp Genotyping Master Mix matching size | Reactions‡ |
|---|---|---|
| 25 μL | 0.5 mL (1 X 0.5 mL) | 100 |
| 250 μL | 5.0 mL (1 X 5 mL) | 1,000 |
| 500 μL | 10 mL (2 X 5 mL) | 2,000 |
| 1250 μL | 25 mL (5 X 5 mL) | 5,000 |
| 2500 μL | 50 mL (1 X 50 mL) | 10,000 |
* For use with real-time qPCR instruments that do not require reference dye.
† For use with real-time qPCR instruments that require reference dye.
‡ Number of reactions based on 10 µL reaction volume.
Table 2. Reference dye requirements for various PCR systems.*
| PCR system | Reference dye required? | |
|---|---|---|
| Yes | No | |
| 7900HT Fast and 7300 Real-Time PCR System (Thermo Fisher Scientific™) | X | |
| StepOne™ and StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific) | X | |
| Mx3005P™ and Mx4000P™ qPCR System (Agilent) | X | |
| 7500 Real-Time PCR System (Thermo Fisher Scientific) | X | |
| ViiA™7 Real-Time PCR System (Thermo Fisher Scientific) | X | |
| QuantStudio™ Flex Systems (Thermo Fisher Scientific) | X | |
| CFX, iQ™, and Opticon™ Real-Time PCR Detection Systems (Bio-Rad) | X | |
| LightCycler® Real-Time PCR Systems (Roche) | X | |
* For instruments not listed, please check with the manufacturer.
Product Data
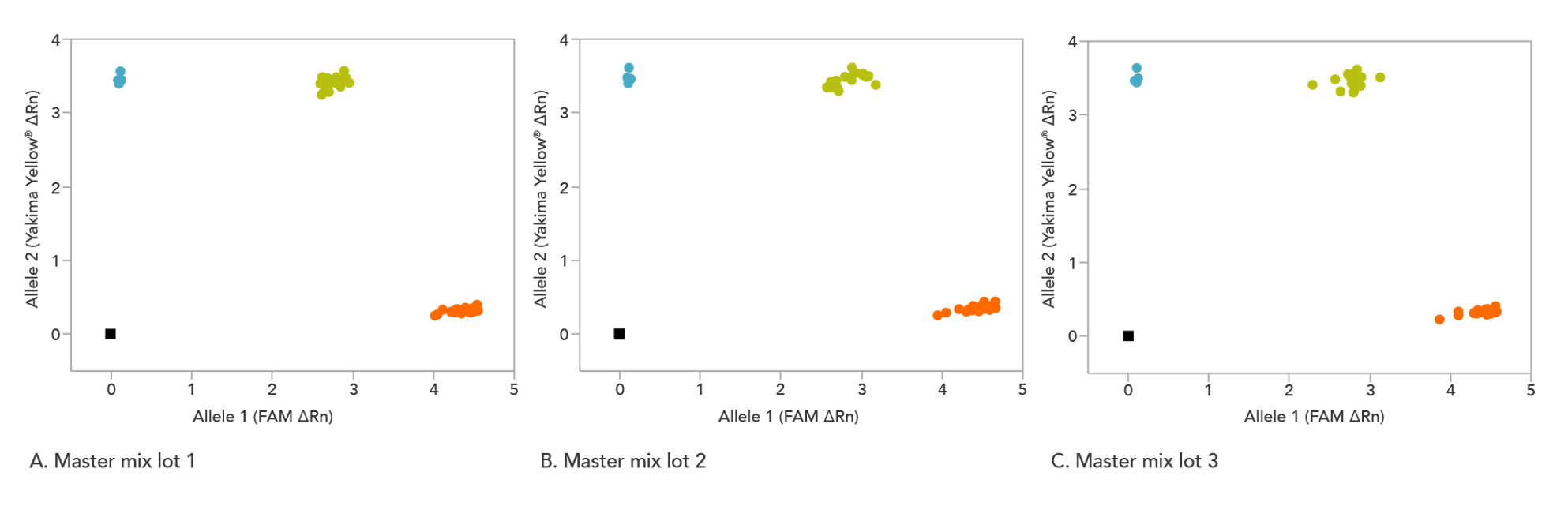
rhAmp Genotyping Master Mix quality assures lot-to-lot consistency
Whether you are just starting with a genotyping study, repeating experiments, or continuing a long-term investigation, the rhAmp Genotyping Master Mix can help you obtain reliable, consistent results (Figure 1).
Figure 2. Consistent genotyping results across multiple lots of rhAmp Genotyping Master Mix. 16 rhAmp SNP Assays were used in 5 µL genotyping reactions, with 3 ng purified genomic DNA samples from 46 individual B cell lines (Coriell Institute) and 3 unique lots of rhAmp Genotyping Master Mix. Genotyping results were analyzed using QuantStudio™ 7 Flex Real Time PCR System software (Thermo Fisher Scientific). The rhAmp Genotyping Master Mix provides consistent results that have been tested to illustrate comparable cluster angles and cluster density with rhAmp SNP Assays. The color of the point represents the homozygous samples with allele 1 (grey), homozygous samples with allele 2 (dark blue), and heterozygous samples with alleles 1 and 2 present (light blue).
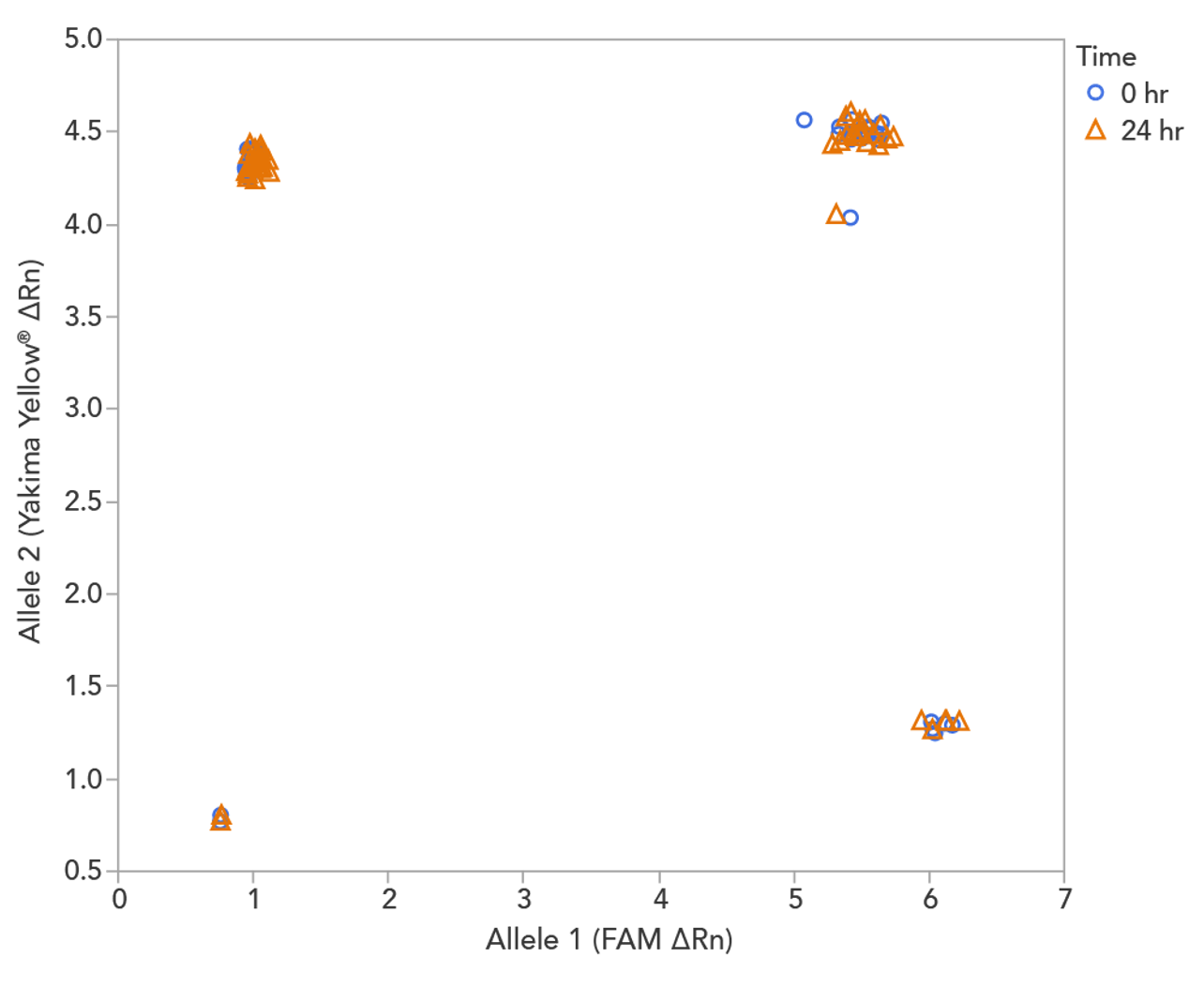
rhAmp SNP Genotyping reaction stability facilitates high‑throughput setup
rhAmp SNP Genotyping reactions are stable for up to 24 hours at room temperature (20–25°C) with all reagents combined before PCR cycling (Figure 2), which facilitates an automated and high-throughput reaction setup.
Figure 3. rhAmp SNP Assays and Master Mix can be used after mixing for up to 24 hr at room temperature. Two identical genotyping reaction plates were prepared, using 16 rhAmp SNP Assays in 5 µL genotyping reactions, with 3 ng of purified genomic DNA samples from 46 individual B cell lines (Coriell Institute). One plate was run immediately at time 0 hr, and the second reaction plate remained on the benchtop for 24 hr before the PCR was performed. Genotyping results were analyzed using QuantStudio 7 Flex Real-Time PCR System software (Thermo Fisher Scientific). The rhAmp Master Mix provides consistent results up to 24 hours after reaction preparation with no major change in genotyping results.
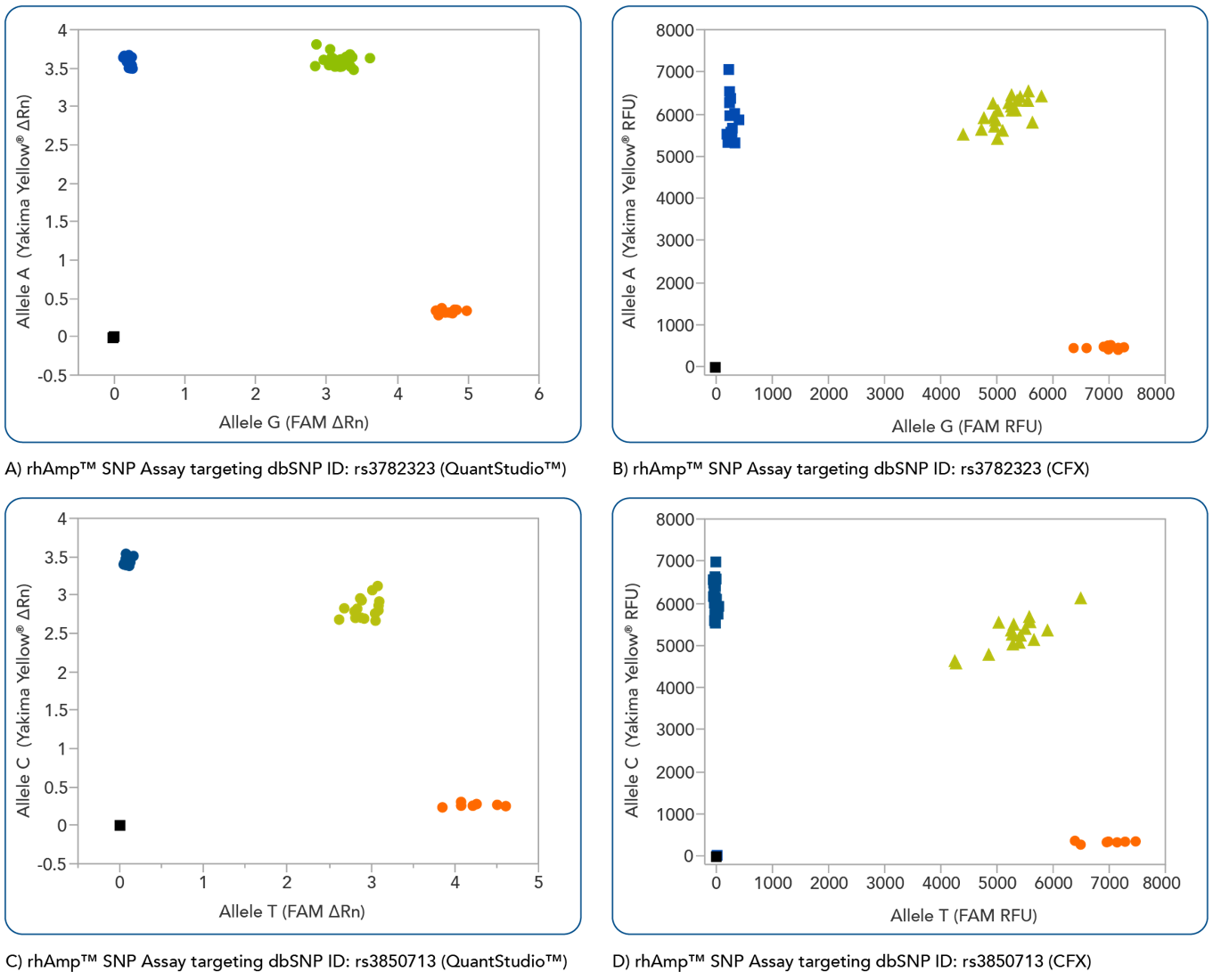
rhAmp SNP Genotyping is compatible with commonly available real-time qPCR platforms
rhAmp SNP Assays are compatible with a variety common real-time qPCR platform (Figure 3). Universal probes are part of the rhAmp Reporter Mix and are available with or without reference dye to ensure compatibility.
Figure 4. rhAmp SNP Assays deliver high precision regardless of qPCR instrument used. Allelic discrimination plots for SNPs located in genes (A and B)
Resources
References
- Beltz KT, D; Wang, J. et al. A High-Performing and Cost-Effective SNP Genotyping Method Using rhPCR and Universal Reporters. Scientific Research. 2018;9(9).