Researchers performing genome editing experiments frequently ask us whether including a crRNA:tracrRNA annealing step is necessary to achieve the highest level of genome editing activity. So, we set up an experiment to test whether CRISPR RNA duplex formation functionally impacts genome editing.
Functional test of RNA annealing step on genome editing
For this experiment we used Alt-R CRISPR-Cas9 System RNAs (see the sidebar, Reagents optimized for CRISPR editing). For the annealing condition, Alt-R crRNAs targeting 2 HPRT sites (HPRT 38285 AS, HPRT 38509 S) were combined with Alt-R tracrRNAs in a 1:1 ratio in either IDTE (10 mM TRIS, pH 7.5; 0.1 mM EDTA) or Duplex Buffer (30 mM HEPES, pH 7.5; 100 mM Potassium Acetate), heated to 95ºC for 5 minutes, and allowed to slowly cool to room temperature. CRISPR RNAs treated with or without this annealing step were reverse transfected (Lipofectamine® RNAiMAX, Thermo Fisher Scientific) into Cas9 expressing HEK293 cells at 10, 20, or 30 nM final concentration. Genomic DNA was isolated 48 hr after transfection, and the HPRT target sites were PCR amplified and digested with T7EI. Resulting fragments were visualized on a Fragment Analyzer™ (Advanced Analytical Technologies).
How important is the crRNA:tracrRNA annealing step?
As seen in Figure 1, including the heating and cooling step to anneal CRISPR RNAs does improve genome editing activity at the lower transfected dose (10 nM). When this annealing step is omitted at this dose, there is a >30% loss in editing activity.
In their paper, Kinetics of the CRISPR-Cas9 effector complex assembly and the role of 3'-terminal segment of guide RNA, Mekler et al provide an explanation for why omitting the crRNA:tracrRNA annealing step results in decreased genome editing activity. They show that the kinetics of RNP formation is ~9-fold slower when crRNA and tracrRNA are not preannealed [1, Fig 6].
While at higher transfection concentrations, the effect is less dramatic, the annealing step is still recommended to maximize genome editing efficiency; it can be particularly helpful for researchers working in sub-optimal or low efficiency settings.
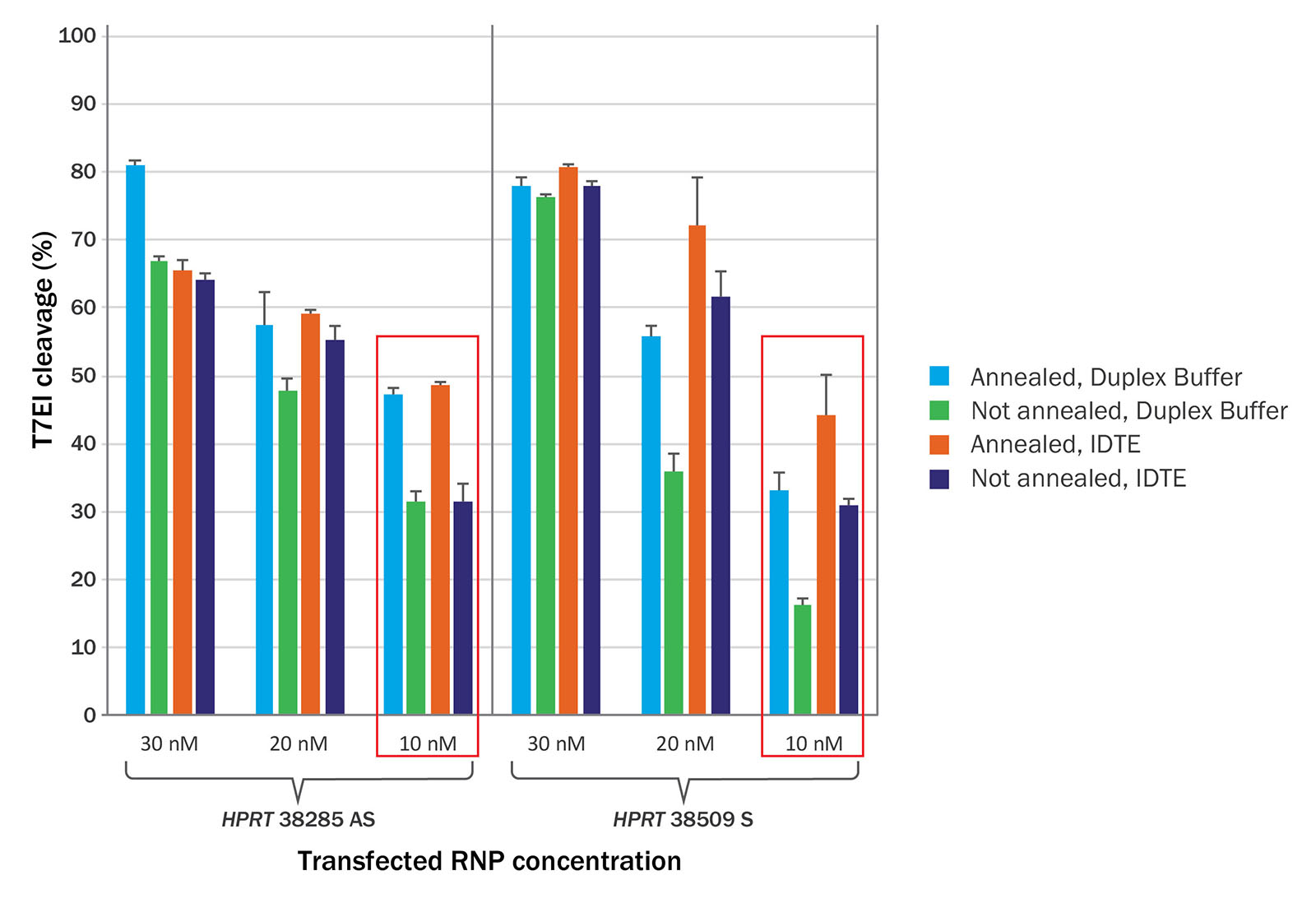
We recommend that users titrate the amount of annealed CRISPR RNA that is transfected into their cells and use the lowest amount in the plateau phase of the dose-response curve (when increasing the amount delivered no longer improves genome editing efficiency) for ongoing experiments. This titration will ensure optimal editing while reducing potential off-target effects and should be done for each new cell line used.
Tested reagents and protocols for optimal genome editing
IDT scientists continue to develop improvements in CRISPR technology. We have demonstrated that you can further optimize genome editing by complexing CRISPR RNAs and Cas9 protein as a ribonucleoprotein (RNP) complex prior to transfection. See the data in the article, Improve your genome editing with the Alt-R S.p. Cas9 Nuclease 3NLS and modified crRNAs. You can also obtain protocols for generating the RNP and deliveito cell lines at www.idtdna.com/CRISPR.
T7EI mismatch endonuclease assay for genome editing analysis
We currently recommend using T7 endonuclease I (T7EI) for CRISPR mutation detection because the method is simple and provides clean electrophoresis results. T7EI analysis is compatible with a broad range of PCR buffers and does not usually require prior purification of the PCR product. Note that T7EI activity is sensitive to DNA:enzyme ratio and incubation temperature and time [2].
T7EI is able to recognize insertions and deletions of ≥2 bases that are generated by non-homologous end-joining activity in CRISPR experiments [3]. Because T7EI does not recognize 1 bp indels, it underrepresents the total editing. For a protocol, see the Alt-R CRISPR-Cas9 System User Guide.

